 Associate Professor
Associate Professor
MSc (Moscow State University, Moscow), PhD (Institute of Microbiology, Moscow)
Principal Investigator, Enzyme Genomics Lab and
BioZone — Centre for Applied Bioscience and Bioengineering
Room: WB412 | Tel.: 416-978-4013 | Email: a.iakounine@utoronto.ca
Memberships
American Association for the Advancement of Science (AAAS)
Research Interests
Enzyme Genomics
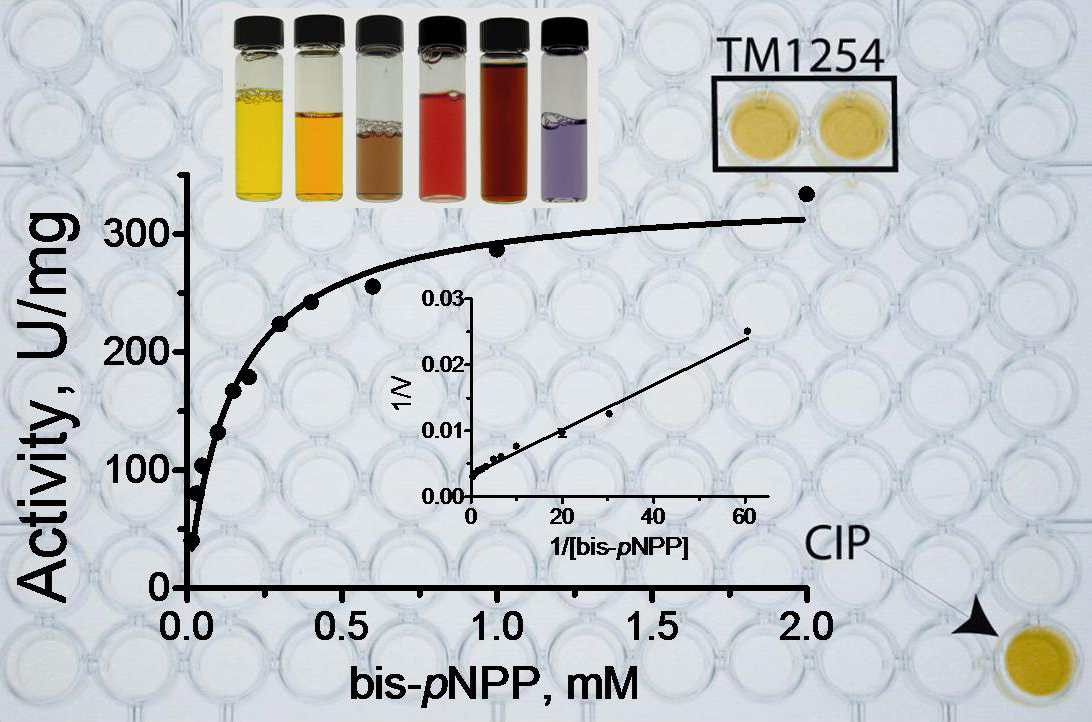
Understanding the function of every protein in the cell is one of the great challenges of the post-genomic era. Global genome sequencing efforts have revealed that in any genome 30-50% of genes encode proteins with unknown function (hypothetical proteins). These proteins need to be experimentally characterized, because their functions can’t be computationally predicted on the basis of sequence homology to known proteins. We are using enzymatic assays to directly test purified unknown proteins for catalytic activity. We have designed a series of general and specific screens to detect hydrolases, oxidoreductases, and transferases. The general screens use either general chromogenic substrates or pools of substrates. The positive hits with the model substrates are then tested with a set of potential natural substrates or the substrate pools are deconvoluted to identify the preferred in vitro substrate. The identification of a biochemical activity of a hypothetical protein helps to determine its cellular role. In a screen of over 3,000 purified proteins, over 750 unknown proteins had clear catalytic activity. Many of them were further characterized using secondary enzymatic screens with specific substrates. Enzymatic screens are also used to quickly characterize large enzyme families and to verify the hypotheses generated by structural studies of proteins with unknown function.
Novel Industrial Enzymes
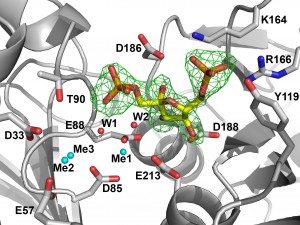
Presently, there is a global drive to promote “white” (or biocatalysis-based) biotechnology as a central element in the transformation of modern industrialized societies to sustainable economies. Despite the obvious benefits of biocatalysis, the major hurdles hindering the application of enzymatic processes are: the high production and enzyme costs, low or moderate yields obtained, the limited repertoire of the available enzymes and their low activity and stability. We are exploring the wealth of sequenced genomes and metagenomes to discover new industrial enzymes for potential applications in bioremediation and bioenergy. Global genome and metagenome sequencing efforts have produced millions of genes encoding unknown proteins. These uncharacterized proteins represent a potentially rich source of novel industrial enzymes. Our approach is based on the high-throughput biochemical and structural characterization of purified predicted hydrolases and oxidoreductases (dehalogenases, cellulases, esterases, lipases, and oxidases). The long-term aim of this project is to discover novel enzymes for applications in biocatalysis, bioenergy and bioremediation.
Molecular mechanisms of CRISPR
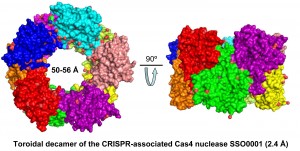
Most prokaryotic genomes contain structures known as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs). Associated with them are a large group of uncharacterized proteins known as CRISPR Associated (Cas). It has been shown that CRISPRs and Cas proteins represent a novel microbial genome defence system which functions to some extent analogously to the eukaryotic RNA silencing machinery. To date, the CRISPR system comprises several thousand uncharacterized proteins organized into approximately 45 protein families and nine types of cas operons (CRISPR-subtypes). Six Cas families (Cas1 to Cas6) represent the core group of CRISPR-associated proteins, and based on the sequence analysis they are predicted to have nuclease and/or helicase activity. We have already biochemically characterized and solved crystal structures of several nucleases from the four core Cas families (Cas1 to cas4). The main goal of our work is to experimentally characterize the biochemical activity and catalytic mechanism of the core Cas proteins and their complexes.
Selected Publications
Nemr, K., Mueller, J., Joo, J.C., Gawand, P., Choudhary, R., Mendonca, B., Lu, S., Yu, X., Yakunin, A.F., and Mahadevan, R.M. Engineering a short, aldolase-based pathway for (R)-1,3-butanediol production in Escherichia coli. Metab. Eng., 48: 13-24, 2018.
Wang, P.-H., Khusnutdinova, A.N., Luo, F., Xiao, J., Nemr, K., Flick, R., Brown, G., Mahadevan, R., Edwards, E.A., and Yakunin, A.F. Biosynthesis and activity of prenylated FMN cofactors. Cell Chem. Biol., 25: 560-570, 2018.
Raj, K., Partow, S., Khusnutdinova, A.N., Yakunin, A.F., and Mahadevan, R. Biocatalytic production of adipic acid from glucose using engineered Saccharomyces cerevisiae. Metab. Eng. Commun., 6: 28-32, 2018.
Khusnutdinova, A.N., Flick, R., Popovic, A., Brown, G., Tchigvintsev, A., Nocek, B., Correia, K., Joo, J.C., Mahadevan, R., and Yakunin, A.F. Exploring microbial carboxylate reductases for the reduction of bifunctional carboxylic acids. Biotechnol. J., 12: 1600751, 2017.
Joo, J.C., Khusnutdinova, A.N., Flick, R., Kim, T., Bornscheuer, U.T., Yakunin, A.F., and Mahadevan, R. Alkene hydrogenation activity of enoate reductases for an environmentally benign biosynthesis of adipic acid. Chem. Sci., 8: 1406-1413, 2017.
Huang, L., Khusnutdinova, A., Nocek, B., Brown, G., Xu., X., Cui, H., Petit, P., Flick, R., Zallot, R., Balmant, K., Ziemak, M.J., Shanklin, J., de Crecy-Lagard, V., Fiehn, O., Gregory III, J.F., Joachimiak, A., Savchenko, A., Yakunin, A.F., and Hanson, A.D. A family of metal-dependent phosphatases implicated in metabolite damage control. Nature Chem. Biol., 12: 621-627, 2016.
Makarova, K.S., Wolf, Y.I., Alkhnbashi, O., Costa, F., Shah, S., Saunders, S.J., Barrangou, R., Brouns, S.J.J., Charpentier, E., Haft, D.H., Horvath, P., Moineau, S., Mojica, F.J.M., Terns, R.M., Terns, M.A., White, M.F., Yakunin, A.F., Garret, R.A., van der Oost, J., Bachofen, R., and Koonin, E.V. An updated evolutionary classification of CRISPR-Cas systems. Nature Rev. Microbiol., 13: 722-736, 2015.
Lemak, S., Beloglazova, N., Nocek, B., Skarina, T., Flick, R., Brown, G., Popovic, A., Joachimiak, A., Savchenko, A., and Yakunin, A.F. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S]-cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J. Am. Chem. Soc., 135: 17476-17487, 2013.
Beloglazova, N., Petit, P., Flick, R., Brown, G., Savchenko, A., and Yakunin, A.F. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J., 30: 4616-4627, 2011.
Babu, M., Beloglazova, N., Flick, R., Graham, C., Skarina, T., Nocek, B., Gagarinova, A., Pogoutse, O., Brown, G., Binkowski, A., Phanse, S., Joachimiak, A., Koonin, E.V., Savchenko, A., Emili, A., Greenblatt, J., Edwards, A.M., and Yakunin, A.F. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol., 79: 484-502, 2011.
Kuznetsova, E., Proudfoot, M., Sanders, S., Reinking, J., Savchenko, A., Arrowsmith, C.H., Edwards, A.M. and Yakunin, A.F. Enzyme genomics: application of general enzymatic screens to discover new enzymes. FEMS Microbiol. Reviews, 29: 263-279, 2005.